Our projects
The main unifiying theme for our lab’s work is understanding how genetics influences our biology and manifests in effects on complex diseases and traits using computational tools and epidemiologic principles.
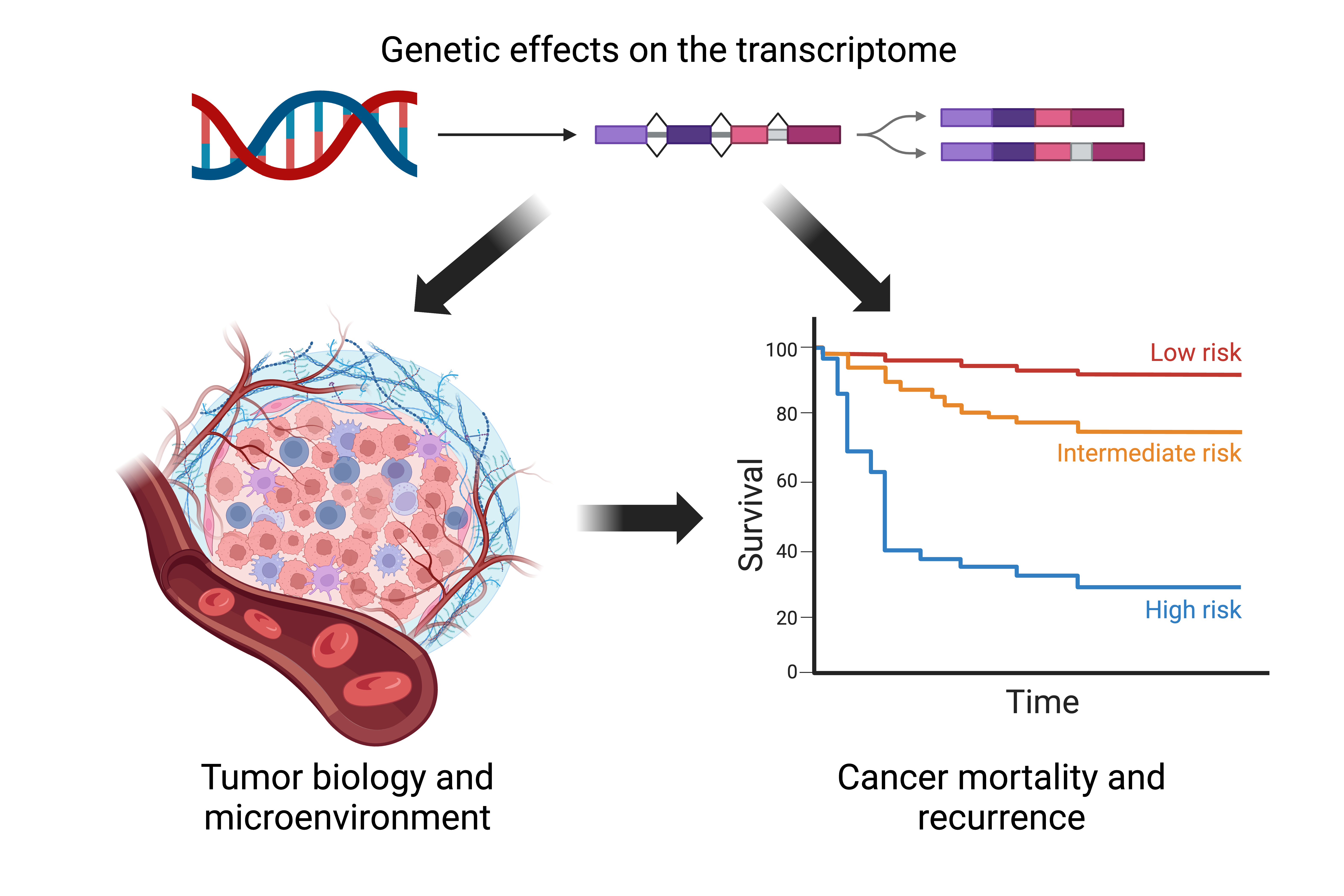
Transcriptomic effects on cancer risk and outcomes
One main goal of our lab is to understand the biological and environmental contributions to cancer risk, outcomes, and disparities. We study the genetic regulation of the transcriptome, how this is modified by the environment, and how this affects cancer risk, mortality, recurrence, and tumor progression and endophenotypes. We focus on how genetic variants influence alternative splicing patterns and isoform expression by analyzing large molecular datasets collected from diverse populations.
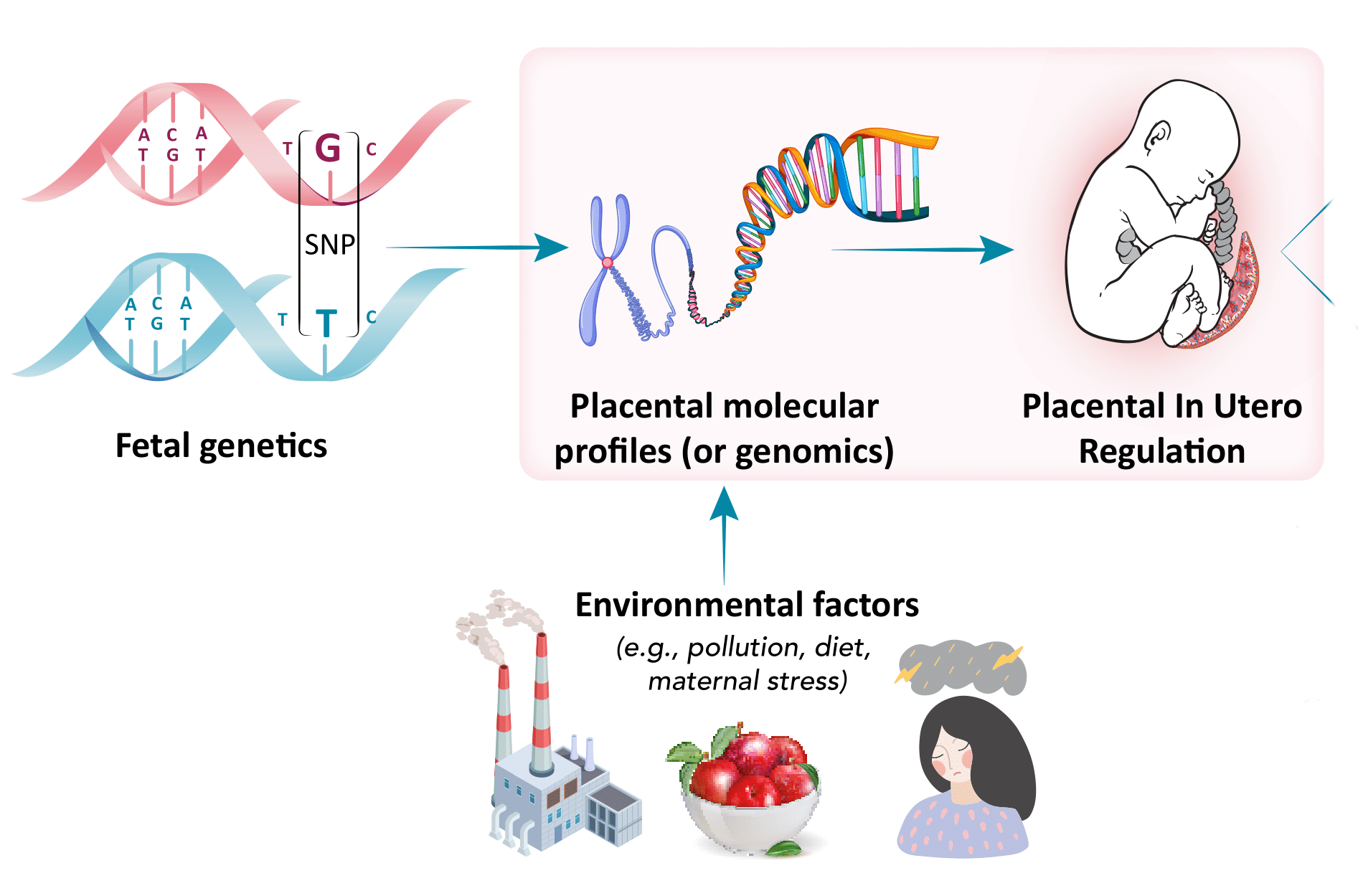
Genetic and epigenetic regulation during development
A second goal of our lab is to generate and analyze large-scale genetic and molecular datasets that can help the research community understand molecular regulation during the developmental window. Our lab focuses on the genomic regulation within the placenta, the master regulator of the intrauterine environment. Through international collaborations, we seek to understand the influence of genetic and epigenetic regulation in the placenta on developmental programming and its effects on early-life outcomes.
To study cancer and development, we build computational tools that use statistical principles and machine learning techniques to integrate genetic and molecular datasets to prioritize biological mechanisms that contextualize genetic associations with complex traits. Here are a few packages we’ve developed.
Featured tools
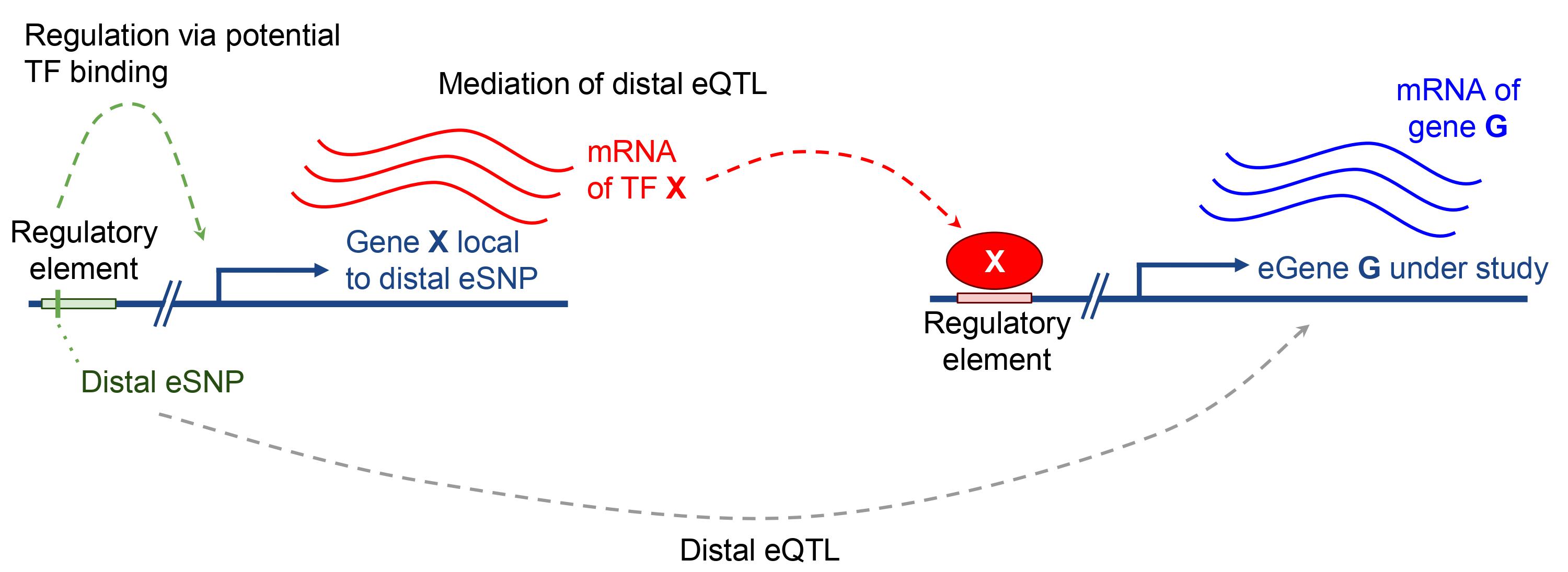
R package to predict gene expression and map gene-trait associations through distal-mediated TWAS
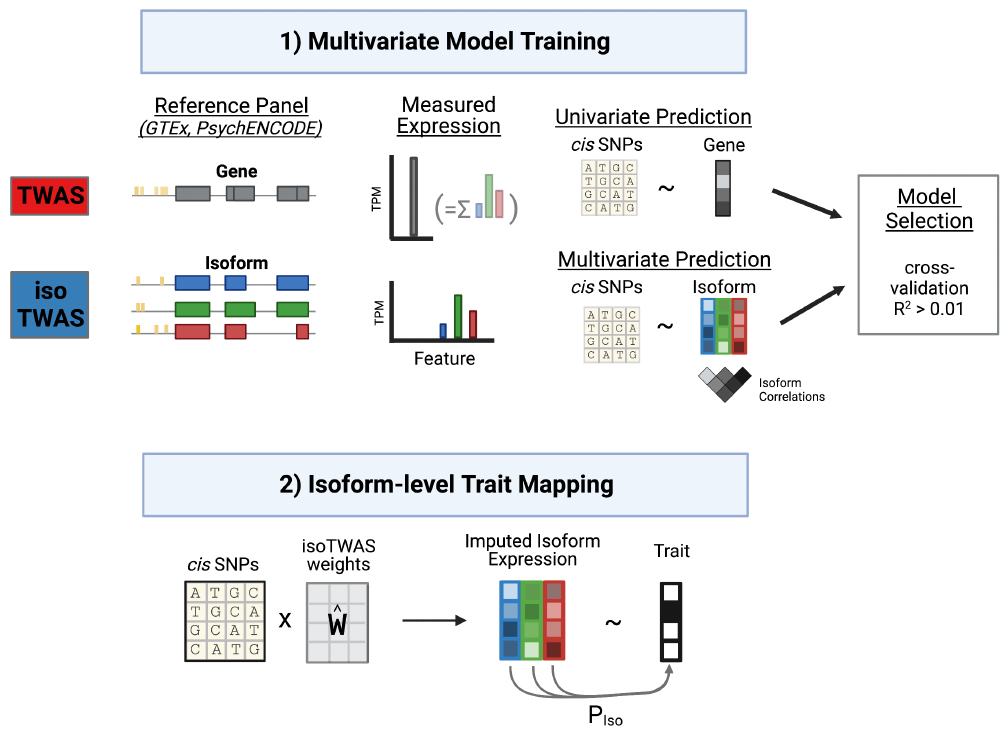
R package to build multivariate isoform models and map isoform-trait associations using two-step associations
More
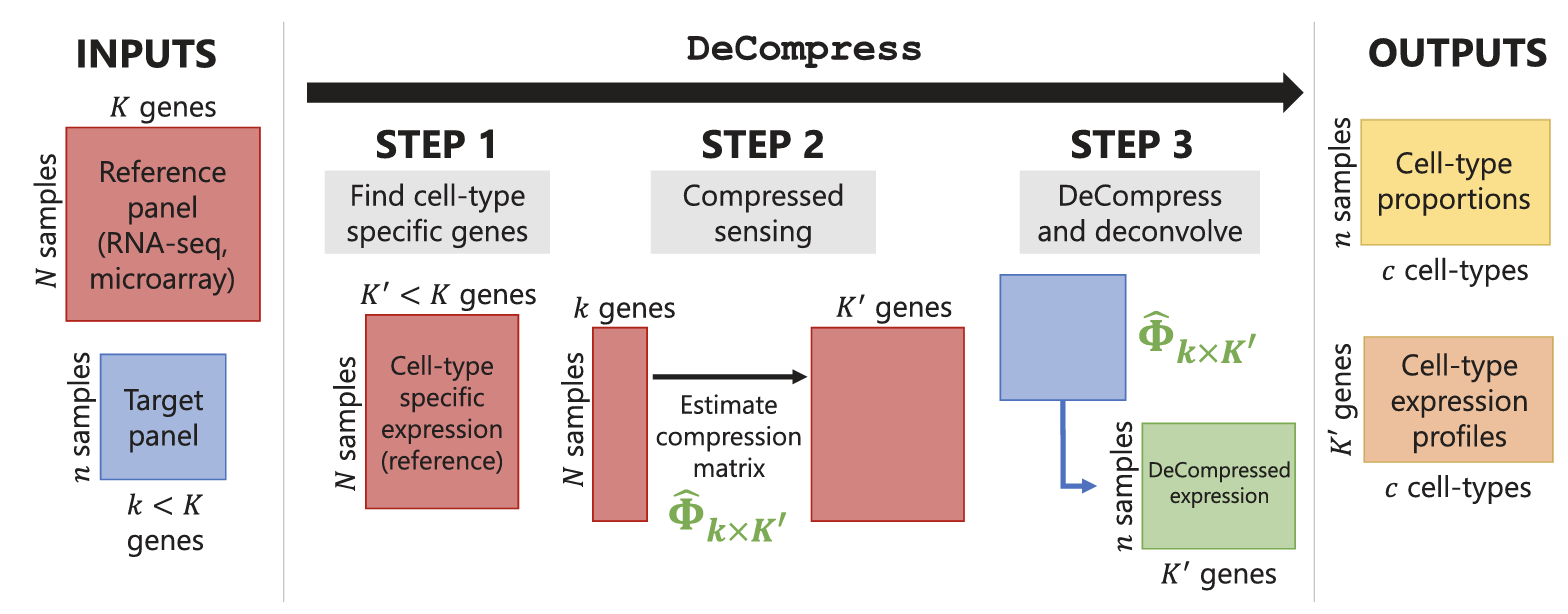
R package to use compressed sensing for cell-type deconvolution for targeted RNA sequencing platforms
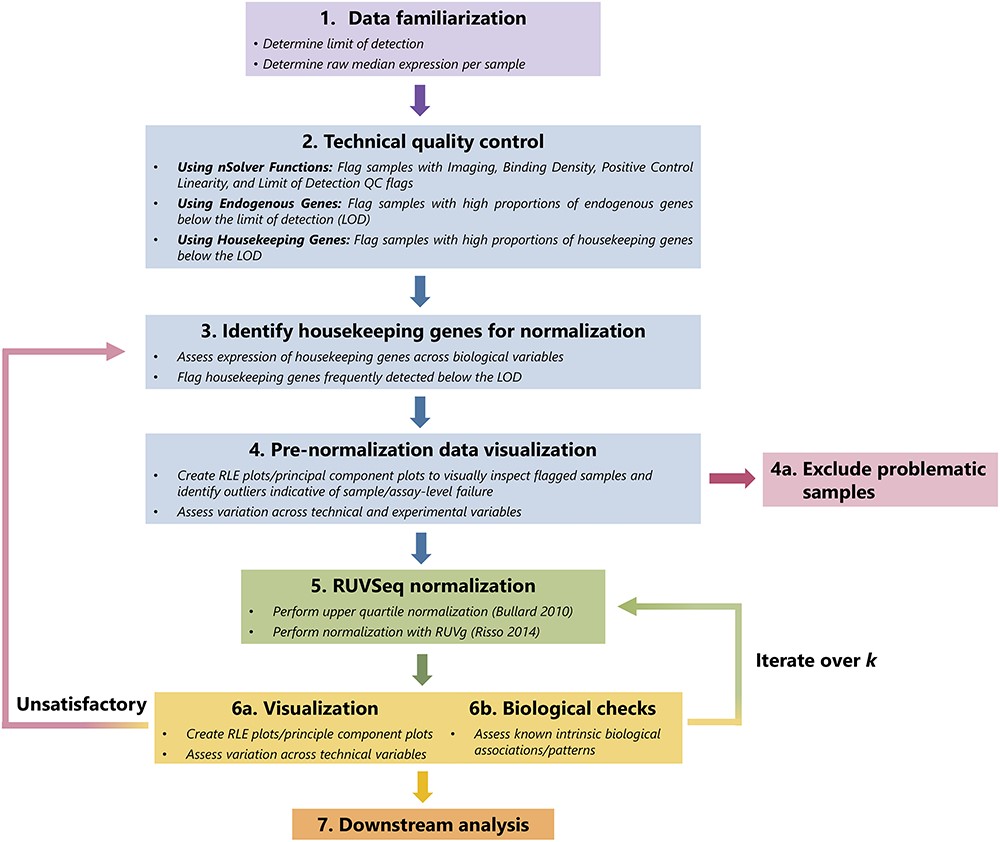
R package to use RUV factor analysis quality control and normalization of NanoString nCounter targeted RNA panels
